Introducing R Markdown for Jupyter with ipymd
Here, I present ipymd, a plugin for jupyter that allows to read and write R Notebook format. The package is freely available from github.
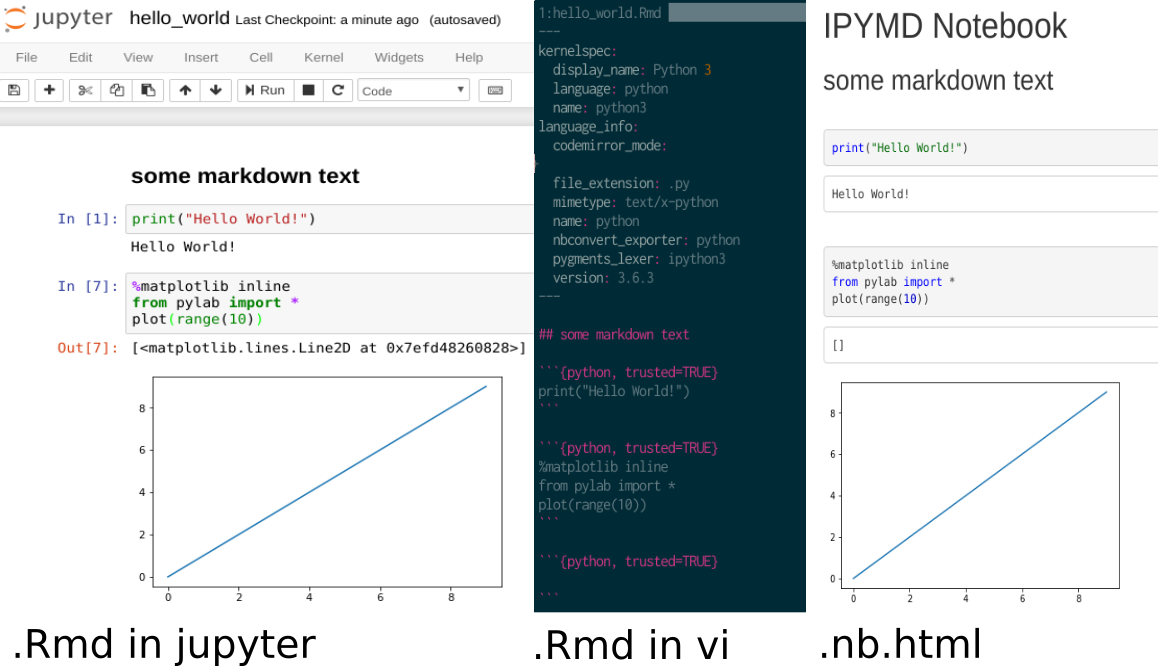
This screenshot shows the same notebook, opened with jupyter, vi and the .nb.html file opened in chrome:
Motivation
Working in Data Science with both R and Python I use both Jupyter notebooks and R Markdown/R Notebooks on a daily basis, depending on the project.
While I usually prefer to work with Python, I have always been envying the R community for the R markdown format due to the following reasons:
- As output is separated from the source, it works well under version control
- The source code can be edited in a text-only editor such as
vi. - The chunk options of R markdown allow to hide certain parts of the document, or to add figure captions, allowing to generate publication-quality reports.
To obtain the best of two worlds, I implemented R markdown (or should it be called Pymarkdown?) support for jupyter.
When it comes to generating publication quality reports, the R markdown/Pymarkdown documents can be fed into Tom Augspurger’s pystitch to obtain a rendered html or pdf report.
How it works
The jupyter plugin is based on rossant/ipymd, an excellent library for converting jupyter notebooks to various formats.
ipymd is a python package that converts on-the-fly between the jupyter notebook-format and the R notebook format. It hooks into jupyter as content-manager, automatically performing such a conversion every time jupyter reads or writes a file, which allows to edit the supported formats directly in jupyter.
The classical jupyter ipynb format is a json-based list of cells, which looks roughly like this:
{
"source": "print('Hello World!')",
"outputs": [{
"text/plain": "Hello World!"
}]
},
{
"source": ...
"outputs": []
}
In RMarkdown, source and output are separated into a .Rmd and .nb.html file respectively.
The former is plain markdown consisting of code chunks:
## Some markdown comments from a jupyter markdown cell
```{python}
print('Hello World!')
```
```{python}
...
The latter is a html file that can readily be viewed in the browser to view the report.
All output is contained in this html document and can be read back by ipymd or RStudio.
